
Home Research Lab Members Publications Education Contact Us Gallery
| 1. Molecular Mechanism of HIV-1 Infection of Resting CD4 T cells |
A major current focus of my laboratory is on the molecular interaction of HIV-1 with human resting CD4 T cells. We are particularly interested in HIV-1 interaction with the chemokine co-receptor CXCR4 in mediating actin cytoskeletal rearrangement required for viral intracellular migration. In the past, our studies have focused on several aspects of virus-CD4 T cell interaction at the cortical actin layer. We studied the effects of viral envelope signaling on the actin rearrangement of resting CD4 T cells. These studies led to the discovery that treatment of resting CD4 T cells with gp120 or HIV-1 triggers the polymerization and depolymerization of filamentous actin (F-actin) in the cortical region. We also identified cofilin as a molecular target of CXCR4 signaling triggered by HIV gp120 to promote the cortical actin dynamics. On the basis of our studies, we proposed a molecular model suggesting a direct involvement of the cortical actin and cofilin in viral latent infection of resting T cells (Figure 1) (1-2). |
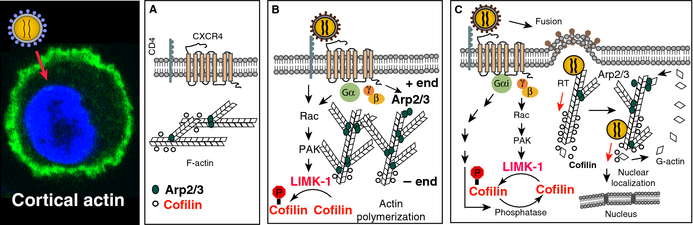 |
Figure 1. Model of chemokine signaling and actin dynamics in HIV infection of CD4+ T cells. (A and B) Engagement of viral gp120 with the receptor/co-receptor on CD4+ T cells results in activation of the Rac1-PAK-LIMK-Cofilin pathway and actin polymerization which transiently block receptor internalization to facilitate fusion. (C) Following entry of the viral core, the pre-integration complex (PIC) undergoes reverse transcription and nuclear migration possibly in association with actin. Nuclear migration is further facilitated by activation of cofilin and the Rac1-WAVE2-Arp2/3 pathway, which promotes actin treadmilling and pushes the viral PIC to the nucleus. |
|
My
laboratory
will
continue to focus on
studying molecular details of how HIV interacts with
the cortical actin
cytoskeleton in human resting CD4 T cells. This is
an
important, yet
largely unexplored research field. Although our
previous studies have
established a role of the cortical actin in HIV
latent
infection of
resting T cells, the molecular mechanisms are
currently not defined.
Mapping these molecular events is fundamental to
establishing a
framework for understanding the early events in
viral
infection of CD4
T cells.
|
| 2. Pre-integration Transcription of HIV-1 DNA |
| In addition to studying HIV interaction with actin, my laboratory is currently also studying pre-integration transcription. In the past, we have demonstrated that pre-integration transcription is a normal process occurring during HIV-1 infection of CD4 T cells and macrophages (3-7). We are in the process of identifying the non-integrated, transcribing HIV DNA templates (Figure 2). |
Figure 2. Pre-integration transcription of HIV-1. Viral transcription in the absence of integration generates all classes of viral transcripts, but only early proteins such as Tat, Rev, and Nef are synthesized at low levels. Tat and Nef can modulate cellular conditions. Viral replication does not occur without integration, but infection by a second virus can rescue the unintegrated viral genomes. |
| 3. Development of a Rev-dependent Lentiviral Vector Targeting HIV-1 Infection |
|
My
laboratory has
developed a
Rev-dependent lentiviral vector to target HIV-infected
macrophages and
T cells, using a bacterial toxin, anthrolysin O from Bacillus
anthracis (8-10).
We are in the process of
constructing a series of Rev-dependent vectors
carrying a variety of
bacterial toxins and human apoptotic genes to induce
the decay of viral
reservoirs in infected macrophages and T cells (Figure
3).
|
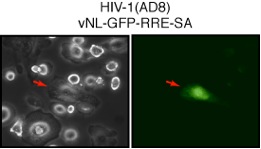 |
Figure 3. Fluorescent microscopy of GFP expression in macrophages infected with HIV-1 and the Rev-dependent GFP lentiviral vector. Cells were infected with HIV-1 (AD8) and then infected with the Rev-dependent lentivirus vNL-GFP-RRE-SA. Infected cells were examined with fluorescent microscopy. The left and right panels show the bright and green fluorescent fields of the same cells. Red arrows indicate an HIV-1-infected cell expressing the GFP-protein. |
|
References
|
___________________________________________________________________________________________________