In
Search of Proline
For this assignment, I first had to pick an amino acid out of the twenty naturally occurring amino acids. The amino acid I chose was proline. Proline is very unique in that it is the only amino acid that has its side chain attached to the amino acid back bone twice, whereas all of the other naturally occurring amino acid side chains are attached only once. Proline is an important amino acid in collagen and cartilage. Collagen is found in tendons, ligaments, and connective tissues in the body. This helps keep joints and tendons cushioned and working smoothly. It also promotes the production of bone, skin, and cartilage. Proline is also effective in preventing wrinkles and the natural aging of skin. Proline is also important in reversing atherosclerotic deposits. It does this in two ways. One is by preventing further build up of atherosclerotic deposits, and the second is to help release “already deposited fat globules from blood vessel wall into the blood serum” [4]. A large source of proline in the human diet is found in meats, eggs, and dairy produces.
Part I:
Choosing the
protein-hetero compound complex
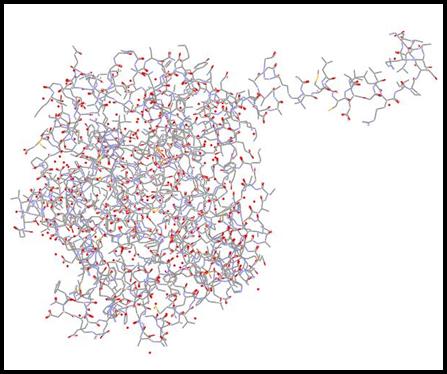 In order to find a protein containing the
heterocompound, proline, I had to search for it on a protein data base. The protein data base that I used was the
RCSB Protein Data Base**, http://www.rcsb.org
is the web address. Here I search by
using the search phrase “proline.” There
were 4,748 articles that were found. I
went through each one that used X-Ray Diffraction and not NMR
spectroscopy. I also looked for the
isolated proline structure as a “Ligand Chemical Component.” The article that I found my protein in was
the fourth article on the third page.
The article is Structure of the E.
coli PutA proline dehydrogenase domain reduced by dithionite and complexed with
proline**. I then downloaded the PDB
file and opened it in DS Visualizer. The
PDB ID Code was 2FZN, and the HET code is PRO.
In order to find a protein containing the
heterocompound, proline, I had to search for it on a protein data base. The protein data base that I used was the
RCSB Protein Data Base**, http://www.rcsb.org
is the web address. Here I search by
using the search phrase “proline.” There
were 4,748 articles that were found. I
went through each one that used X-Ray Diffraction and not NMR
spectroscopy. I also looked for the
isolated proline structure as a “Ligand Chemical Component.” The article that I found my protein in was
the fourth article on the third page.
The article is Structure of the E.
coli PutA proline dehydrogenase domain reduced by dithionite and complexed with
proline**. I then downloaded the PDB
file and opened it in DS Visualizer. The
PDB ID Code was 2FZN, and the HET code is PRO.
![Text Box: Figure 1: The protein chain containing the heterocompound found in the article from the RCSB Protein Data Base [2]](Protein-Modelinga_files/image036.gif)

Figure 2: The
Cleaned up Protein structure containing the
heterocompound
proline, which is circled in purple.**[2]
Once in DS Visualizer, I cleaned up the protein. I hid the water molecules that were associated with it. I also hid the second compound chain that was associated with the proline molecule. I displayed the protein as a solid ribbon, along with making the proline structure a ball and stick molecule so that it could be seen easier.

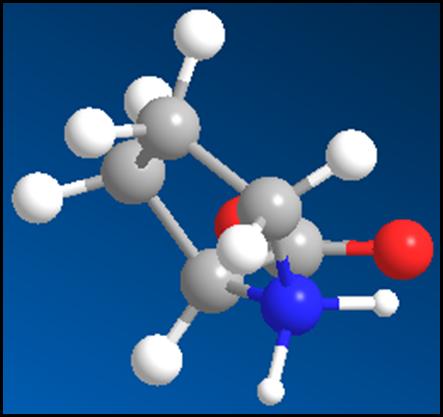
Figure 3: This is the extracted
proline molecule that
has the charges corrected on it
so that it is a zwitterions
molecule with a positive charge on the nitrogen and a
negative charge on the
oxygen.**[2]
I then extracted the proline molecule from the protein by copying and pasting it into a new window of DS Visualizer. Here, I added hydrogen atoms which gave the molecule an incorrect structure and formula. The program made the nitrogen atom neutral and turned one of the oxygen atoms into an alcohol. I gave the carboxyl group a negative charge but removing the hydrogen, and I added a hydrogen to the nitrogen atom so that it would have a positive charge. I then saved the image in three different formats, . mol. .jpg, and .msv.
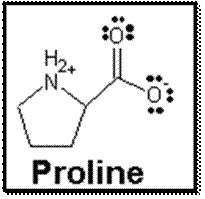
Figure 4: This is the proline
structure
drawn in the program ChemSketch,
with the appropriate charges, unpaired
electrons, and
stereochemistry showing.
Finally, I drew
the proline molecule in the program ChemSketch.
Here I drew the basic structure of the molecule, including the
stereochemistry by adding the wedge where appropriate. I also added a positive charge to the
nitrogen by adding a hydrogen atom to it, along with giving the oxygen atom a
negative charge by removing the hydrogen.
I showed the unpaired electrons to the oxygen atoms as well. I then save the image as a .sk2 file because
I was unable to save it as a .gif and reopen it.
Part II:
Displaying the Protein Hetero-Compound Complex
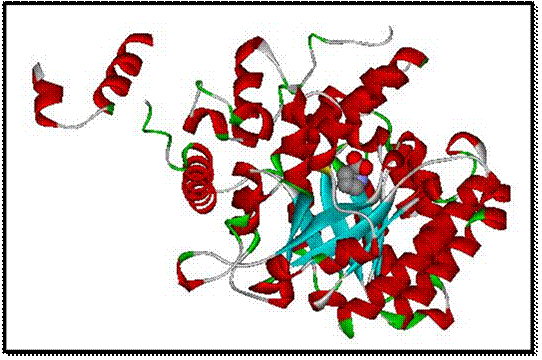
Figure 5: This is the secondary protein structure
with the heterocompound, proline, in the center in the CPK format. **[1]
I made
the protein image appear in the secondary structure form as a solid
ribbon. I also put the heterocompound,
proline, in the space filling CPK format.
Part III:
Steric Energy Calculations

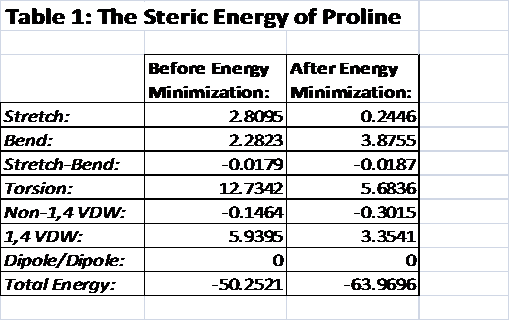
Figure 6: The Heterocompound,
Proline complexed with the protein, before energy minimization.[1]
There have been two procedures performed here. First, I took a single point energy calculation. With this molecular position, the heterocompound, proline, is in the angle that it would be when it is complexed with the protein. In the single point energy calculation, the stretch, bend, stretch-bend, torsion, Non-1,4 van der Waals, 1,4 van der Waals, dipole/dipole interactions, and the total energy was calculated. The second single point energy calculation was performed after the heterocompound, proline, was twisted into the structure that has the lowest amount of energy.
Stretch and Bend and Stretch-Bend:
The Stretch and bend energies make a significant contribution to the total energy of the molecule. The stretch energy was calculated to be 2.8095 and the bend energy was 2.2823. The stretch-bend energy had very little impact on the total energy because it was so low, which is also true for the minimized structure as well. They were both below -0.019. The stretch energy for the minimized structure was 0.2446, which is much lower than the complexed structure. The stretch energy barely made a contribution to the total energy because it was so small. The bend energy for the minimized structure did have a significant amount of energy, at 3.8755. It actually had a greater amount of energy than in the complexed structure before it was minimized at 2.2823.
Torsion:
The torsion is the angel between the main atoms in the proline molecule. This would include the oxygen atom, the nitrogen atom, and the five carbon atoms in the ring. This measures the amount of energy in the strained angles of proline. There is an extreme difference between the torsion of the complexed structure and minimized structure. The torsion for the complexed structure was calculated to be 12.7342, whereas the minimized structure calculated it to be 5.6836. The difference is over seven. The complexed structure clearly was getting more energy from the torsion between its major atoms. The minimized structure was obviously in an optimized position to have as little torsion as possible in its total energy use.
Figure 7: The heterocompound, Proline, after the
energy minimization. [1]
Non-1,4 VDW and 1,4 VDW:
For both the minimized and the complexed structures, the Non-1,4 van der Waals interaction calculations were determined to be very low and negative. This is because the interactions are between atoms that are more than three bonds away from one another. For this molecule, these interactions are typically between hydrogen atoms, which explain why these interactions are so low. The 1,4 van der Waals interactions are very different from the non-1,4 van der Waals interactions. These have a positive and a higher energy because these are the interactions of atoms that are within three bonds of each other. It makes sense that these would have a greater impact on the total energy of the molecule.
Dipole/Dipole interactions:
For both of the structures, there was no dipole/dipole interaction between the atoms so it didn’t contribute to the total energy of the molecule.
Total Energy:
Over all the total energy for the complexed molecule is much higher than the minimized structure. As a complexed structure, the atoms in the structure are strained and are producing a lot of energy. Once minimized, the structure is less energetic because it is at the optimized position. Though neither of the structures have a large amount of total energy, there is still a major difference between the two.
Superimposing
the Extracted and the Energy Minimized 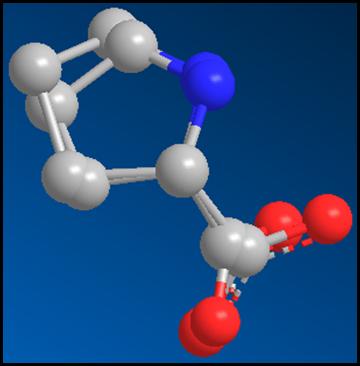 Hetero Compounds
Hetero Compounds
Figure 8: These
are the two proline structures
before and
after the energy
minimization, overlaid
with one another. [1]
In order to obtain the overlay of these two proline structures, I copied them into the same window in Chem3D. Here, I selected one fragment and used the “select target fragment” in the overlay functions to select the second fragment. I then overlaid the two structures together. It is very important to hide the hydrogen, mainly because they do not really overlay with one another and it makes it easier to see the main atoms like the nitrogen atom, the carbon atoms, and the oxygen atoms.
It is clear to see that the “goodness of fit” for these two structures is not perfect. This is to be expected, since one structure is the molecule while it is complexed with the protein and the other is in the minimized energy structure. Only the second carbon bonding the ring to the carboxyl group was lined up well with the other one. The part of the ring that specifically did not overlay well would be the end with the carbons that were the farthest from the carboxyl group. When proline is complexed with the protein, the ring is bent back, whereas with the minimized energy structure, it is brought forward to put less strain on the ring. Also, the oxygen atoms in the carboxyl group appear to be in different plans. In the structure complexed with the protein, the oxygen atoms are out of the plan of the ring and rotated. In the energy minimized structure, the oxygen atoms are more so in the plain of the ring and are brought forward more. The bending of the structure of proline complexed with the protein is most likely due to the interactions of the other molecules and forces in the protein structure itself, whereas the second structure is optimized for the least amount of energy when it is in the gaseous phase and not interaction with other molecules. These can be seen in a different way below, in Figure 9, which is the structure of proline complexed with the protein, and in Figure 10, which is the structure of proline after the energy minimization.
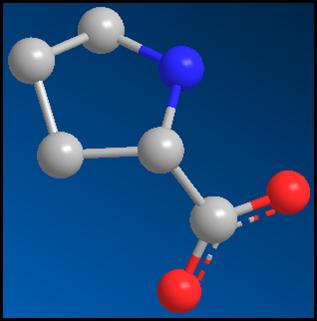
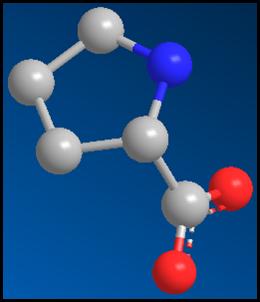 Figure 9:
Figure 9:
The structure
of proline
complexed with
the protein Figure
10:
before energy
minimization has occurred. The
structure of proline after the energy
minimization has occurred.
Part IV:
Protein-Ligand
Interactions
I used the search data base, PDBsum, to find more specific information on my protein containing my heterocompound. The search engine gave a wire diagram, as seen to the left. This diagram is an illustration in a linear format that describes the interactions between the amino acid residues in the protein. The “spiral” like sections is referring to the α-helixes, whereas the multiple β are referring to the β-sheets throughout the protein. Also, the small red dots that are located above some of the residues is in reference to the residue coming into contact with the ligand, proline. This diagram gives a more numerical and linear way of seeing the interactions. Within this protein, there are multiple residues interacting with the ligand.
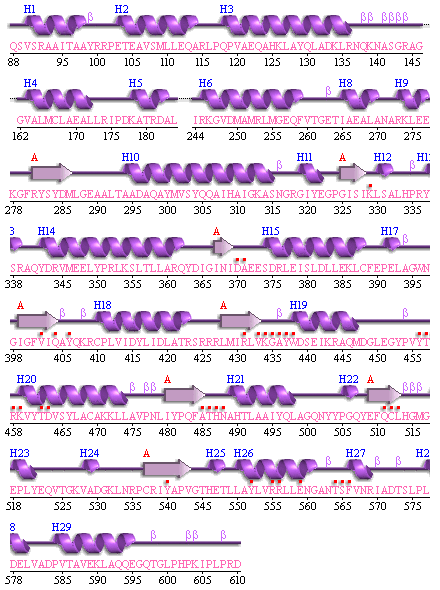
Figure 11: This
is the wiring diagram of the protein that contains the proline molecule. From
the PDBsum website***[3]
 Figure 12: This is the
LIGPLOT of the interactions between proline and other amino acids within the
protein. From PDBsum website***[3]
Figure 12: This is the
LIGPLOT of the interactions between proline and other amino acids within the
protein. From PDBsum website***[3]
Above is a diagram of a LIGPLOT obtained from the same PDBsum website. A LIGPLOT is a plot of the main amino acid residues that are interacting with the ligand, proline. In this diagram, it shows that there are three main molecules that are interacting with the proline molecule by hydrogen bonding. These three main molecules are lysine329, two different Arginine molecules, arginine556 and Arginine 555. These three molecules are interacting with the carboxyl group of the proline molecule. These are not the only molecule interacting with the proline of course. There is also alaine436, tyrosine552, tyrosine540, aspartic acid370, and leucine513. Also, the other ligand, Fad2001 is also interacting with the proline. This diagram helps clarify which molecules bond how and where to the proline and what is really interacting with it.
Figure 13: The protein structure with proline as a ball and stick in the
center.

I
began the process of displaying the interactions of the residues with the
proline ligand in the three-dementional format.
I did this to see if the 3-D format showed the same interactions as the
2-D LIGPLOT does. I did this using DS
Visualizer. I started with the protein
being displayed at a solid ribbon and the proline molecule as a ball and stick.
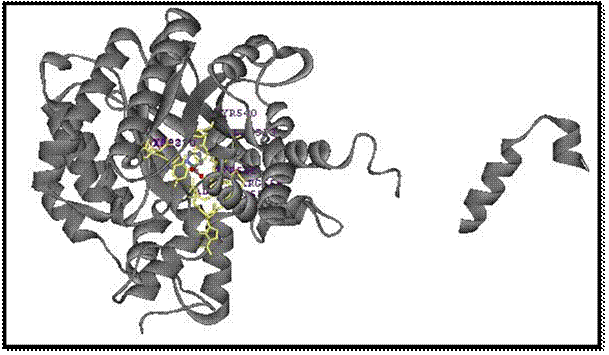 Figure 14: The
protein structure displayed as a solid ribbon,
Figure 14: The
protein structure displayed as a solid ribbon,
with the proline heterocompound in
the center as a
ball and stick, and the
interactions between the
surrounding residues with the
ligands.
The
next major step is to display all of the residues that are interacting with the
proline molecule. I selected all of the
residues that were listed in the LIGPLOT diagram above. I made these a pale yellow color in the stick
formation with their labels. This
illustrates where the residues are located in the protein in relation to the
proline. It is obvious that the
molecules are relatively close in proximity to the proline molecule.
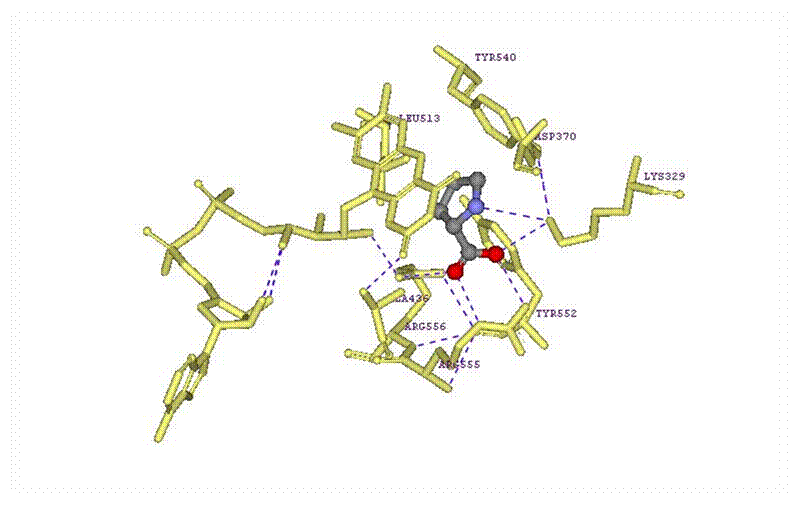 Figure 15: A diagram of the proline heterocompound with the
interactions of the surrounding amino acid residues.
Figure 15: A diagram of the proline heterocompound with the
interactions of the surrounding amino acid residues.
Here is a better illustration of the interactions between the proline molecule and the other amino acid residues. For the most part, all of the interactions among the residues were the same as shown in the LIGPLOT diagram above. However, some of the placement of the residues was not completely accurate, but that is mainly because the LIGPLOT is a 2-D diagram. What is interesting to see though is that there is an interaction between the nitrogen atom and the lysine molecule which was not shown in the LIGPLOT. There are also interactions between the other residues with one another. This was also not shown in the LIGPLOT.
 Figure 16: Again, here is an
image, like the previous one, that shows the protein as a solid ribbon, with
Figure 16: Again, here is an
image, like the previous one, that shows the protein as a solid ribbon, with
the proline
molecule as a ball and stick, and the interacting amino residues in the pale
yellow stick format.
***The wiring diagram and the LIGPLOT diagram both came from
the PDBsum website: Laskowski R A,
Chistyakov V V, Thornton J M (2005). PDBsum more: new summaries and
analyses of the known 3D structures of proteins and nucleic acids. Nucleic
Acids Res., 33, D266-D268.
Date visited May 1, 2008. http://www.ebi.ac.uk/thornton-srv/databases/cgi-bin/pdbsum/GetPage.pl?pdbcode=2fzn&template=main.html*.***
Part V:
Protein-Ligand
Interactions
Table 2: Interactions between
Proline and other Residues
|
Amino acid
residue |
Hetero
compound atoms |
Nature of
interaction |
|
|
Lysine 329 |
Side chain – NH2+ is positively charged. |
The oxygen of the carboxyl group |
This is a hydrogen interaction. The nitrogen is the Hydrogen Bond Donor (HBD). |
|
Asparagine 370 |
side chain is neg. charged carboxylate |
???? |
It doesn’t seem to be close to Proline. Asp370 is interacting with Lys329 and Tyr540 by hydrogen bonding. |
|
Alanine 436 |
Side chain – methyl group |
????? |
It is hard to say because this residue is not close to proline. However, it is interacting with the other heterocompound FAD 2001 by van der Waals interactions. |
|
Leucine 513 |
side chain – CH3-CH2-CH3 |
Pyrrolidine ring |
It is hard to say. Probably a van der Waals interaction between one of the methyl groups of Leu513 and the pyrrolidine ring of proline. |
|
Tyrosine540 |
side chain -- - CH2Ph-OH |
Carboxyl group |
It is hard to say. Probably hydrogen interactions – the Tyr540 is the hydrogen bond donor (HBD) whereas the Proline is the hydrogen bond acceptor (HBA). |
|
Tyrosine552 |
side chain - CH2Ph-OH |
????? |
Tryosine552 doesn’t seem to be in close contact. No hydrogen bond interactions to other residues. Possibly electrostatic interactions with one of the nitrogen groups on Arginine555. |
|
Arginine 555 |
side chain – nitrogen groups |
The carboxyl group |
Electrostatic interactions – the positively charged nitrogen is interaction with the double bonded oxygen of the carboxyl group. The nitrogen in the chain is also interacting with the O- of the carboxyl group. |
|
Arginine 556 |
side chain – nitrogen groups |
The O- of the carboxyl group. |
Electrostatic interactions – the two positively charged nitrogen groups on the Arginine and interacting with the negatively charged oxygen atom of the Carboxyl group on Proline. |
|
FAD 2001 |
side chain |
Possibly the pyrrolidine ring |
It is unclear what the interactions are between FAD2001 and proline. It could be van der Waals interactions between the carbon groups of FAD2001 and proline, or it could be the pi-stacking interactions between the pyrrolidine ring of the proline and the pyridine rings of FAD 2001. |
Bibliography:
[1] H.M. Berman, J. Westbrook, Z. Feng, G. Gilliland, T.N. Bhat, H. Weissig, I.N. Shindyalov, P.E. Bourne: The Protein Data Bank. Nucleic Acids Research 28 pp. 235-242 (2000) Data Visited: April 19, 2008. http://www.rcsb.org/pdb/explore.do?structureId=2FZN. DOI 10.2210/pdb2fzn/pdb
[2] W. Zhang, M. Zhang, W. Zhu, S. Wanduragula, D. Rewinkel, J.J. Tanner, D.F. Becker. “Structure of the E. coli PutA proline dyhydrogenase domain reduced by dithionite and complexed with proline.” 2006. Date Visited April 19, 2008.*** http://www.rcsb.org/pdb/explore.do?structureId=2FZN. DOI 10.2210/pdb2fzn/pdb
[3] Laskowski R A, Chistyakov V V, Thornton
J M (2005). PDBsum more: new summaries and analyses of the known 3D structures
of proteins and nucleic acids. Nucleic Acids Res., 33,
D266-D268. Date visited May 1,
2008. http://www.ebi.ac.uk/thornton-srv/databases/cgi-bin/pdbsum/GetPage.pl?pdbcode=2fzn&template=main.html.
[4] HealthyVitaminsGuide.com. “Proline”.
HealthyVitaminsGuide.com. http://www.healthvitaminsguide.com/aminoacids/proline.htm . Data visited May 6, 2008.
[5] Vitaminstuff.com. “Proline.”
VitaminStuff 2007. http://www.vitaminstuff.com/amino-acid-proline.html. Date visited May 6, 2008.
***I could not
find an article that had an abstract that also had proline as a ligand chemical
component. I searched each article
within the first twenty pages the search found.***